To apply machine learning algorithms to generate models capable of predicting mortality in COVID-19 patients, using a large platform of plasma inflammatory mediators.
DesingProspective, descriptive, cohort study.
Setting6 intensive care units in 2 hospitals in Southern Brazil.
PatientsPatients aged > 18 years who were diagnosed with COVID-19 through reverse transcriptase reaction or rapid antigen test.
InterventionsNone.
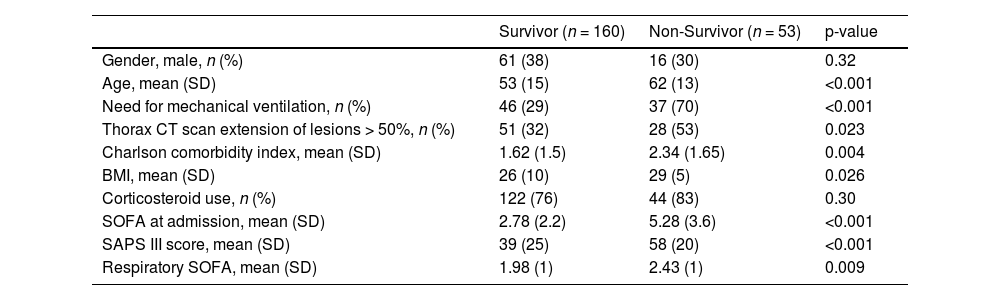
Main variables of interestDemographic and clinical variables, 65 inflammatory biomarkers, mortality.
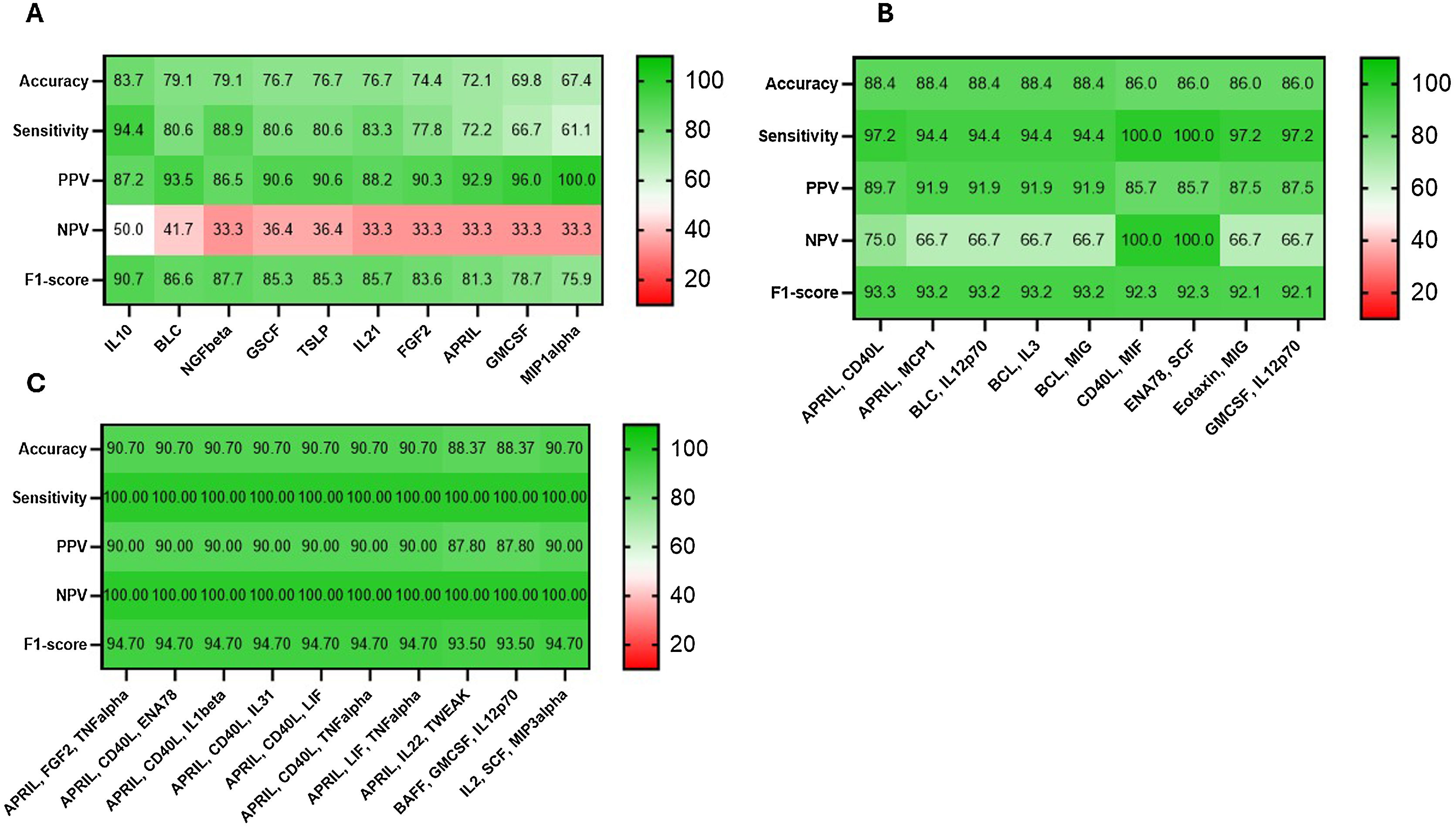
ResultsCombinations of two or three proteins yield higher predictive value when compared to individual proteins or the full set of the 65 proteins. A proliferation-inducing ligand (APRIL) and cluster of differentiation 40 ligand (CD40L) consistently emerge among the highest-ranking combinations, suggesting a potential synergistic effect in predicting clinical outcomes. The network structure suggested a dysregulated immune response in non-survivors characterized by the failure of regulatory cytokines to temper an overwhelming inflammatory reaction.
ConclusionOur results highlight the value of feature selection and careful consideration of biomarker combinations to improve prediction accuracy in COVID-19 patients.
Aplicar algoritmos de aprendizaje automático para generar modelos capaces de predecir la mortalidad en pacientes con COVID-19, utilizando una amplia plataforma de mediadores inflamatorios plasmáticos.
DiseñoEstudio de cohorte prospectivo, descriptivo.
Ámbito6 unidades de cuidados intensivos en 2 hospitales del sur de Brasil.
PacientesPacientes mayores de 18 años diagnosticados con COVID-19 mediante reacción de transcriptasa inversa o prueba rápida de antígeno.
IntervencionesNinguna.
Variables de interés principalesVariables demográficas y clínicas, 65 biomarcadores inflamatorios, mortalidad.
ResultadosLas combinaciones de dos o tres proteínas muestran un mayor valor predictivo en comparación con proteínas individuales o el conjunto completo de las 65 proteínas. APRIL y CD40 L consistentemente aparecen entre las combinaciones de mayor rango, lo que sugiere un posible efecto sinérgico en la predicción de los resultados clínicos. La estructura de la red sugiere una respuesta inmunitaria desregulada en los no sobrevivientes, caracterizada por la incapacidad de las citocinas regulatorias para moderar una reacción inflamatoria abrumadora.
ConclusiónNuestros resultados destacan la importancia de la selección de características y la cuidadosa consideración de combinaciones de biomarcadores para mejorar la precisión predictiva en pacientes con COVID-19.
Article
Go to the members area of the website of the SEMICYUC (www.semicyuc.org )and click the link to the magazine.