Sepsis is a major public health problem and a leading cause of death in the world, where delay in the beginning of treatment, along with clinical guidelines non-adherence have been proved to be associated with higher mortality. Machine Learning is increasingly being adopted in developing innovative Clinical Decision Support Systems in many areas of medicine, showing a great potential for automatic prediction of diverse patient conditions, as well as assistance in clinical decision making. In this context, this work conducts a narrative review to provide an overview of how specific Machine Learning techniques can be used to improve sepsis management, discussing the main tasks addressed, the most popular methods and techniques, as well as the obtained results, in terms of both intelligent system accuracy and clinical outcomes improvement.
La sepsis representa un problema de salud pública de primer orden y es una de las principales causas de muerte a nivel mundial. El retraso en el inicio del tratamiento, junto con la no adherencia a las guías de práctica clínica se asocian a una mayor mortalidad. El aprendizaje automático o machine learning están siendo empleados en el desarrollo de sistemas de apoyo a la decisión clínica, innovadores en muchas áreas de la medicina, mostrando un gran potencial para la predicción de diversas condiciones del paciente, así como en la asistencia durante el proceso de toma de decisiones médicas. En este sentido, este trabajo lleva a cabo una revisión narrativa para proporcionar una visión general de cómo las técnicas de machine learning pueden ser empleadas para mejorar el manejo de la sepsis, discutiendo las principales tareas que tratan de resolver, los métodos y las técnicas más empleados, así como los resultados obtenidos, tanto en términos de precisión de los sistemas inteligentes, como en la mejora de los resultados clínicos.
Sepsis is a major public health problem and a leading cause of death in the world. Although it is not easy to reliably measure incidence and mortality,1–4 most of the recent data shows both an increase in incidence and number of deaths and a decrease in case-fatality.
In addition to the difficulty in obtaining reliable data, the sepsis definition, first established in 1991 (Sepsis-1), has been updated in 2001 (Sepsis-2) and 2016 (Sepsis-3),5–7 which makes temporal comparison difficult. A meta-analysis that includes 22 studies published in high-income countries between 1979 and 2015 shows an incidence of 288 hospital-treated sepsis and 148 hospital-treated severe sepsis per 100,000 person-year.8 When only studies of the last decade are analyzed, incidence increases to 437 hospital-treated sepsis and 270 hospital-treated severe sepsis per 100,000 person-year. Hospital mortality during this period was 17% for sepsis and 26% for severe sepsis.
From a complementary perspective, in Spain the incidence and mortality data are very heterogeneous. Incidence is nearly 100 cases per 100,000 person-year9 and hospital mortality ranges from 43% to under 20%.9–12
To sum up, although the numbers vary, data seem to confirm that incidence and number of deaths due to sepsis increases but the case-fatality decreases.
In a separate but complementary perspective, it is also necessary to consider the consumption of health resources. In this regard, it has been published that mean cost per severe sepsis episode is around $20,000.13,14
Different measures have shown a beneficial impact in terms of reducing mortality, and together with improvement in Intensive Care Unit (ICU) assistance, are the cause of this reduction. To improve survival of septic patients, these measures must be applied as soon as possible. There are three basic mainstays in sepsis management15,16: (i) early administration of adequate antimicrobial therapy,17,18 (ii) resuscitation with fluids and vasopressors,19 and (iii) source control.20 Sepsis bundles have been the cornerstone of the improvement of the quality of sepsis care since 2005. Nowadays, hour-1 sepsis bundle includes five measures that must be accomplished in the first hour since the suspicion of sepsis,21 underlining the importance of time in sepsis treatment.
Despite the fact that adherence with management guidelines has been related with mortality reduction in several studies, sepsis bundles compliance is low.22–24 In this context, different approaches have been tried to enhance guidelines compliance. As an example, educational interventions can achieve a temporary improvement in bundles compliance and, even, a reduction in hospital mortality, but this impact is usually transitory.25
Other types of interventions, such as the design of detection and management programmes for sepsis at the hospital or state level26,27 show similar results and seem to be cost-effective.13
Recently, a new approach to the problem of sepsis has arisen, based on the application of new information technologies. These initiatives range from relatively simple systems of automatic detection of sepsis, using electronic medical record data28,29 or computerized protocols,30 to more sophisticated systems based on Big Data and Artificial Intelligence (AI) designed to detect and even predict sepsis or guide clinical decisions.
Concretely, Machine Learning (ML), a subfield of AI, has gained attention in the sector of medicine. ML goes beyond classic “expert systems”, whose rules are manually coded into them, by creating a new generation of systems built by “learning” from big amounts of data and by dealing with a high number variables simultaneously in order to mimic or even improve human clinical decision making.31 There are applications of ML to almost all medical fields. Some recent reviews of the use of ML in several medical areas have been published, including medical image analysis,32 cardiovascular medicine,33 in critical care,34 or neuro oncology.35
ObjectivesThe main objective of this work is to conduct a narrative review to provide an overview of how specific ML techniques can be used to improve sepsis management, focusing on the following specific tasks: detection, prediction of sepsis/shock, mortality prediction, hospital stay, costs and adherence to guides. Non-AI or management systems for sepsis treatment, as well as non ML-based expert systems, fall outside the scope of this paper and have not being included in the bibliographic research done.
This review has been designed with the aim of being useful, mainly, for clinicians wanting to know how ML could help them in their daily practice, but also for researchers conducting their own studies applying ML to any sepsis treatment related task. Keeping this in mind, five main questions of interest for clinicians have been defined during the design phase and answered in this paper after a broad bibliographic research. For those readers with no background in ML, a brief explanation of the main ML techniques employed in the papers found during the bibliographic research was included. In addition, the technical information of the studies covered in this review, such as the frequency at which each ML technique was used to solve different tasks, the size and design of the studies or the performance results, may help researchers to design their own approaches in the field.
MethodologyWe conducted broad queries through different search engines, including PubMed, Google Scholar, ResearchGate, and ScienceDirect, using the following keywords: sepsis, machine learning, early, prediction, severe sepsis, mortality, detection, artificial intelligence, and data mining. The last search was conducted on November 7, 2019.
Titles and abstracts of the initially gathered contributions were checked to exclude those papers that fall outside the scope of this study, such as studies related to neonatal sepsis. After that, 34 papers remained from international conferences and renowned journals published in the last 13 years (2007-November 2019). From the careful reading of the selected contributions, three main tasks were initially identified as the fundamental objective related to sepsis management: (i) sepsis detection, focused on the identification of septic patients, (ii) sepsis prediction, focused on determining which patients are in risk of developing sepsis, and (iii) mortality prediction, focused on determining which septic patients are in risk of death. Table 1 summarizes the type techniques proposed in the reviewed papers for dealing with these tasks.
Main tasks comprising sepsis management with reference to ML approaches used by the reviewed studies.
| Task | ML technique | References |
|---|---|---|
| Sepsis detection | Artificial Neural Networks | (36) (37) (38) |
| Bayesian | (39) (40) | |
| Decision Trees | (39) (41) (42) (43) (44) (45) | |
| Logistic Regressions | (46) (47) | |
| Support Vector Machines | (39) (48) (49) (50) | |
| Sepsis prediction | Artificial Neural Networks | (37) (38) (51) (52) (53) (54) |
| Decision Trees | (41) (55) (56) (45) (57) | |
| Other Techniques | (55) (58) (51) | |
| Mortality prediction | Artificial Neural Networks | (59) (60) |
| Bayesian | (61) (62) | |
| Decision Trees | (63) (64) | |
| Logistic Regressions | (65) (59) (63) | |
| Support Vector Machines | (59) (61) (66) (64) | |
| Other techniques | (67) (64) | |
Supplementary material S1 provides a detailed view of the main features that characterize all the included studies, while Tables 2–4 contain information about the ML techniques and features used on each study and the performance results achieved for the different tasks.
Main characteristics of the ML classifiers used for sepsis detection.
| Ref | ML type | Variables | Patients | AUC (CI 95%) | Sensitivity (Sen) | Specificity (Sp) |
|---|---|---|---|---|---|---|
| Mao et al., 201842 | GTB | 6 (SaO2, HR, SBP, DBP, T°, and RR) | ED, hospital wards and ICU patients | Sepsis: 0.92 (0.90–0.93)Severe sepsis: 0.87 (0.86–0.88)Septic Shock: 0.9992 (0.9991–0.9994) | (Sen fixed near 0.80):Sepsis: 0.95 (0.93–0.97)Severe sepsis: 0.85 (0.84–0.86)Septic shock: 0.9990 (0.9987–0.9993) | (Sp fixed near 0.80):Sepsis: 0.98 (0.96–1.00)Severe sepsis: 0.996 (0.989–1.00)Septic shock: 1.00 (1.00–1.00) |
| Gonçalves et al., 201339 | DTNBSVM | 9 (bilirubin, creatinine, glucose, leukocytes, platelets, HR, MBP, SBP, and T°) | ICU | DT: 1.00NB: 0.9982SVM: 1.00 | DT: 1.00NB: 1.00SVM: 1.00 | DT: 1.00NB: 0.9990SVM: 1.00 |
| Futoma et al., 201738 | MGP-RNN (LSTM) | 77 (34 physiological variables (6 vital signs, 28 laboratory values),35 covariates (29 comorbidities and other 6), and 8 medication classes) | Hospital | >0.90 | ||
| Horng et al., 201748 | SVM | 12 (age, gender, acuity, SBP, DBP, HR, Pain Scale, RR, SaO2, T°, free text chief complaint, and free text nursing assessments) | ED patients | Vitals: 0.67CC: 0.83BoW: 0.86Topics: 0.85 | Vitals: 0.56CC: 0.75BoW: 0.78Topics: 0.80 | Vitals: 0.68CC: 0.75BoW: 0.79Topics: 0.75 |
| Tang et al., 201049 | SVM | 3 (Non-invasive cardiovascular variables: ECG, Fin-PPG, and Ear-PPG) | ED patients with SIRS | Severe sepsis: 0.78 | 0.9444 | 0.6250 |
| Faisal et al., 201847 | LR | 19 (First electronically recorded vital signs and blood test results) | ED patients | Sepsis: 0.7908Severe sepsis: 0.9036 | Sepsis: 0.5434Severe sepsis:0.5306 | |
| Wang X. et al., 201836 | KELM | 5 (d-xylose, acetatic acid, linoleic acid, d-glucopyranosiduronic acid, and cholesterol) | ED and ICU patients | 0.8957 | 0.6577 | |
| Nachimuthu et al., 201240 | DBN | 10 (WBC, % of immature neutrophiles, HR, MBP, DBP, SBP, T°, RR, PaCO2, and age) | ED patients | First 3h: 0.91102First 6h: 0.91499First 12h: 0.93362First 24h: 0.94353 | First 3h: 0.68902First 6h: 0.70732First 12h: 0.81707First 24h: 0.85976 | First 3h: 0.94881First 6h: 0.94994First 12h: 0.94881First 24h: 0.94539 |
| Desautels et al., 201641 | GTB | 8 (age, HR, DBP, SBP, T°, RR, SaO2, and GCS) | ICU | 0.88±0.006 | 0.80 | 0.80 |
| Kam, H.J. and Kim, H.Y., 201737 | DFNLSTM | 9 (pH, SaO2, WBC, HR, SBP, PP, T°, RR, and age) | SIRS criteria (hospital and ICU) | LSTM: 0.99 | LSTM: 0.97 | LSTM: 1.00 |
| Back et al., 201646 | LR | 7 (HR, DBP, T°, RR, age, admission via ED, LOS) | Hospital | Test/Validation0.96/0.95 | Test/Validation0.96/0.77 | Test/Validation0.83/0.96 |
| Arvind et al., 201850 | SVM | Clinical notes | ICU post-surgical patients | 0.82 | 0.79 | 0.85 |
| Delahanty et al. 201943 | GTB | 13 (8 laboratory results, 3 vital signs, 2 “engineered”) | ED patients | Time after an index timea:0.93 at 1h0.95 at 3h0.96 at 6h0.97 at 12h0.97 at 24h | Time after an index timea:0.68 at 1h0.72 at 3h0.75 at 6h0.79 at 12h0.85 at 24h | Time after an index timea:0.96 at 1h0.97 at 3h0.97 at 6h0.96 at 12h0.96 at 24h |
| Calvert et al., 201944 | GTB | 6 (DBP, SBP, HR, T°, RR, SpO2) | High-risk patients (age≥45 years and length-of-stay≥4 days) | 0.917 | 0.799 | 0.860 |
| Barton et al., 201945 | GTB | 6 (SaO2, HR, SBP, DBP, T°, and RR) | Hospital, ICU, ED patients | 0.88 | 0.80 | 0.78 |
Table's acronyms and abbreviations:
AUC: area under curve; CI: confidence interval; BoW: bag of words; CC: classifier chains; AUROC: area under the receiver operating characteristic curve; DBN: dynamic Bayesian network; DFN: deep feedforward networks; GTB: gradient tree boosting; DT: decision tree; LR: logistic regression; LSTM: long short term memory; KELM: kernel extreme learning machine; ML: machine learning; MGP: multiple-output gaussian process; NB: naïve bayes; RNN: recurrent neural network; SVM: support vector machines.
DBP: diastolic blood pressure; Ear-PPG: ear photoplethysmography; ECG: electrocardiogram, ED: emergency department; ICU: intensive care unit; SIRS: systemic inflammatory response syndrome; Fin-PPG: finger photoplethysmography; GCS: glasgow coma scale; HR: heart rate; LOS: length of stay; MBP: mean blood pressure; PaCO2: partial pressure of carbon dioxide; PP: pulse pressure; RR: respiratory rate; SaO2: oxygen arterial saturation; SBP: systolic blood pressure; T°: temperature; WBC: white blood cells.
NS: no specified.
Index time.
Main characteristics of the ML classifiers used for sepsis prediction.
| Ref | ML type | Variables | Objectives | Patients | AUC (CI 95%) | Sensitivity (Sen) | Specificity (Sp) |
|---|---|---|---|---|---|---|---|
| Desautels et al., 201641 | GTB | 8 (age, HR, DBP, SBP, T°, RR, SaO2, and GCS) | Sepsis prediction | ICU | 4h before sepsis: 0.74±0.010 | 0.80 | 0.54 |
| Futoma et al., 201738 | MGP-RNN (LSTM) | 77 (34 physiological variables, 35 covariates and 8 medication classes) | Sepsis prediction | Hospital | 2h before: ∼0.874h before:∼0.846h before: ∼0.8212h before: ∼0.77 | ||
| Kam, H.J. and Kim, H.Y., 201737 | DFNLSTM | 9 (pH, SaO2, WBC, HR, SBP, PP, T°, RR, and age) | Sepsis prediction | SIRS criteria (hospital and ICU) | LSTM 1h before: 0.96LSTM 2h before: 0.94LSTM 3h before: 0.929DFN100 3h before: 0.915 | LSTM 1h before: 0.92LSTM 2h before: 0.89LSTM 3h before: 0.914DFN100 3h before: 0.886 | LSTM 1h before: 1.00LSTM 2h before: 1.00LSTM 3h before: 0.944DFN100 3h before: 0.944 |
| Wyk et al., 201857 | RF | 8 (HR, RR, SBP, DBP, T°, SpO2, WBC, LOS) | Sepsis prediction | ICU patients | 1h before: 0.7 | 1h before: 0.8 | 1h before: 0.6 |
| Nemati et al., 201858 | Weilbull-Cox Hazards Model | 65 (30 laboratory values, 6 high-resolution dynamical features, 10 clinical features, 19 demographics/context features) | Sepsis prediction | ICU patients | 4h before: 0.846h before: 0.828h before: 0.8212h before: 0.79 | Fixed at 0.85 | 4h before: 0.646h before: 0.628h before: 0.6212h before: 0.57 |
| Wang, R.Z. et al., 201855 | LRSVMLMT | 12 (P, Ca, Mg, BUN, Hb, Platelets, WBC, INR, Alkaline Phosphatase, HR, SBP, and SaO2) | Sepsis prediction | ICU patients | 6h before:LR: 0.685SVM: 0.674LMT: 0.750 | 6h before:LR: 0.752SVM: 0.566LMT: 0.671 | 6h before:LR: 0.618SVM: 0.783LMT: 0.830 |
| Giannini et al.201956 | RF | 587 (demographics, vital signs and laboratory results) | Sepsis prediction | Hospital wards | 0.88 | 0.26 | 0.98 |
| Schamoni. et al., 201951 | LiRMLP | 57 (42 vital signs and laboratory results, 3 demographic, 10 pre-existing conditions) | Sepsis prediction | ICU patients | LiR:0.808 (0.786–0.830) 12–8h before sepsis onset0.770 (0.739–0.801) 24–12hMLP:0.817 (0.789–0.844) 12–8h before0.776 (0.739–0.811) 24–12h before | ||
| Barton et al., 201945 | GTB | 6 (SaO2, HR, SBP, DBP, T°, and RR) | Sepsis prediction | Hospital, ICU, ED patients | 0.84 24h before sepsis onset0.83 48h before sepsis onset | 0.80 24h before sepsis onset0.84 48h before sepsis onset | 0.72 24h before sepsis onset0.66 48h before sepsis onset |
| Scherpf et al., 201952 | RNN | 10 (age, DBP, SBP, pH, SaO2, T°, HR, RR, PaCO2, WBC) | Sepsis prediction | ICU patients | 3h before: 0.81 (0.78–0.84). | (Sp fixed: 0.90)3h before: 47.06h before: 44.912h before: 46.3 | (Sen fixed at 0.90)47.0 (95% CI: 43.1%–50.8%) |
| Kaji et al., 201953 | LSTMRNN | 119 (demographic data, vitals, labs, and treatment) | Sepsis Prediction | ICU patients | Same-day sepsis: 0.952Next-day sepsis: 0.876 | Same-day sepsis: 0.73Next-day sepsis: 0.57 | |
| Fagerström et al., 201954 | LSTM | 24 (demographic data, vital signs, laboratory results, treatment) | Sepsis prediction | ICU patients | 0.83 (48h before) | ||
| Mao et al., 201842 | GTB | 6 (SaO2, HR, SBP, DBP, T°, and RR) | Sepsis severity prediction | Septic patients in ED, hospital wards and ICU | 4h beforeSevere sepsis: 0.85 (0.79–0.91)Septic shock: 0.96 (0.94–0.98) | ||
| Lin et al., 201869 | LSTM | 43 (6 vital signs, 11 laboratory values, 4 treatment, 18 culture results and 4 other) | Sepsis severity prediction | Hospital patients with suspected infection | 12h before septic shock: 0.9411 | 12h before septic shock: 0.8408 | |
| Liu et al., 201970 | GLMXGBoostRNN | 32 (13 vital signs, 12 laboratory results, 7 treatment) | Septic shock prediction | ICU patients with suspected infection | GLM: 0.82XGBoost: 0.83RNN: 0.85 | GLM: 0.85XGBoost: 0.77RNN: 0.79 | GLM: 0.73XGBoost: 0.73RNN: 0.77 |
| Median early warning time:GLM: 9.5hXGBoost: 9.0hRNN: 10.3h | |||||||
Table's acronyms and abbreviations:
AUC: area under curve; CI: confidence interval; DFN: deep feedforward networks; GTB: gradient tree boosting; LiR: linear regression; LR: logistic regression; LMT: logistic model tree; LSTM: long short term memory; GLM: generalized linear model; XGBoost: extreme gradient boosting; ML: machine learning; MGP: multiple-output gaussian process; MLP: multilayer perceptron; RF: random forest; RNN: recurrent neural network; SVM: support vector machines.
BUN: blood urea nitrogen; Ca: Calcium; DBP: diastolic blood pressure; Ear-PPG: ear photoplethysmography; ED: emergency department; ICU: intensive care unit; SIRS: systemic inflammatory response syndrome; GCS: glasgow coma scale; Hb: haemoglobin; HR: heart rate; INR: international normalized ratio; LOS: length of stay; Mg: magnesium; MBP: mean blood pressure; P: phosphorus; PaCO2: partial pressure of carbon dioxide; PP: pulse pressure; RR: respiratory rate; SaO2: oxygen arterial saturation; SBP: systolic blood pressure; T°: temperature; WBC: white blood cells.
NS: no specified.
Main characteristics of the ML classifiers used for mortality prediction.
| Ref | ML type | Variables | Patients | AUC (CI 95%) | Sensitivity (Sen) | Specificity (Sp) |
|---|---|---|---|---|---|---|
| Ribas et al., 201265 | LR | 34 (demographic, comorbidities, organ function, treatment, infection). First 24h of evolution | Severe sepsis admitted to ICU | 0.75 | 0.64 | 0.84 |
| Gultepe et al., 201461 | NBSVM | 5 (Lactate, MBP, RR, T°, and WBC) | Hospital patients with SIRS | NB: 0.660±0.050SVM: 0.726±0.045 | NB: 0.879SVM: 0.949 | NB: 0.385SVM: 0.308 |
| Tsoukalas et al., 201566 | SVM | 5 (T°, RR, WBC, MBP, and lactate) | Patients with SIRS | 0.61 | ||
| Taylor et al., 201663 | RFCARTLR | >500 (demographic, previous health status, ED health status, ED services render, and operational details) | ED septic patients | 28 days mortalityRF: 0.860 (0.819–0.900)CART: 0.693 (0.620–0.766)LR: 0.755 (0.689–0.821) | ||
| Byrne et al., 201659 | LRMLPSVM | 939 peptides from LC-MS/MS at<16h and 48h after septic shock diagnosis. | ICU patients with septic shock | <16h after shock diagnosis:LR: 0.7415MLP: 0.7222SVM: 0.839448h after shock diagnosis:LR: 0.9710MLP: 0.9928SVM: 1.0000 | <16h after shock diagnosis:LR: 0.5556MLP: 0.4444SVM: 0.722248h after shock diagnosis:LR: 1.0000MLP: 1.000SVM: 1.0000 | <16h after shock diagnosis:LR: 0.9275MLP: 1.0000SVM: 0.956548h after shock diagnosis:LR: 0.9420MLP: 0.9855SVM: 1.0000 |
| Wang T. et al., 201862 | DBN | 24 (age, 8 vital signs, 12 laboratory test, GCS, 2 treatments) | Infected ICU-patients | 0.913 (0.906–0.919) | 0.825 (0.802–0.849) | 0.874 (0.802–0.849) |
| García-Gallo et al., 201867 | LASSOSGB | 47 (14 laboratory tests, 9 vital signs, 4 data taken at the time of ICU admission, 14 comorbidities, and 6 organ dysfunction) | ICU patients with sepsis | LASSO: 0.792 (0.791–0.793)SGB: 0.8039 (0.8033–0.8045) | ||
| Chiew et al., 201964 | kNNRFABGTBSVM | 28 (6 vital signs, 22 HRV parameters) | ED patients | kNN: 0.06RF: 0.56AB: 0.38GTB: 0.50SVM: 0.63 | ||
| Perng et al., 201960 | RFkNNSVMSoftMaxCombined with three feature extraction methods:CNNAEPCA | 53 (demographic data, vital signs, laboratory results) obtained during ED stay | ED patients | 72h mortality – Best Classifier:CNN+SoftMax: 0.9428 days mortality – Best Classifier:CNN+SoftMax: 0.92 |
Table's acronyms and abbreviations:
AUC: area under curve; CI: confidence interval; AB: adaptive boosting; AE: auto encoder; CART: classification and regression tree; CNN: convolutional neural network; DBN: dynamic Bayesian network; GTB: gradient tree boosting; LASSO: least absolute shrinkage and selection operator; SGB: stochastic gradient boosting; LR: logistic regression; kNN: k-nearest neighbours; ML: machine learning; MLP: multilayer perceptron; NB: naïve bayes; PCA: principal component analysis; RF: random forest; SVM: support vector machines.
Ca: calcium; DBP: diastolic blood pressure; Ear-PPG: ear photoplethysmography; ED: emergency department; ICU: intensive care unit; SIRS: systemic inflammatory response syndrome; GCS: glasgow coma scale; HRV: heart rate variability; MBP: mean blood pressure; RR: respiratory rate; T°: temperature; WBC: white blood cells.
Based on the study of the aforementioned papers, the following section introduces the main ML concepts and algorithms used in these papers. This section is followed by five sections that try to answer key questions specifically related to actual challenges present in sepsis management. The information provided is of special relevance to researchers who want to contribute advances in the area.
Machine learning overviewML is a discipline of the AI field focused on making machines able to do tasks without being explicitly programmed for them. To do so, algorithms need to be trained, which, depending on the algorithm, can be done by analyzing sample, or training, data or by iteratively developing a strategy for solving problems based on rewards or punishments. Among the different types of learning strategies, one of the most widely used is supervised learning. Here, the objective is to create a model able to predict some output value given a set of input variables.
For example, to predict the malignant (vs. benign) condition of a given tissue given some morphological variables, patient data or even the specimen's image. If the output variable is limited to a known set of values, the task is called classification, whereas if the prediction should be any numerical value within a range, the objective is to do a regression.68 In supervised learning, algorithms need a set of resolved samples, that is, data including the input variables values along with the correct output. Once trained, these algorithms can make predictions over new samples.
Following, we briefly explain the main supervised ML algorithms used in the studies reviewed in this work.
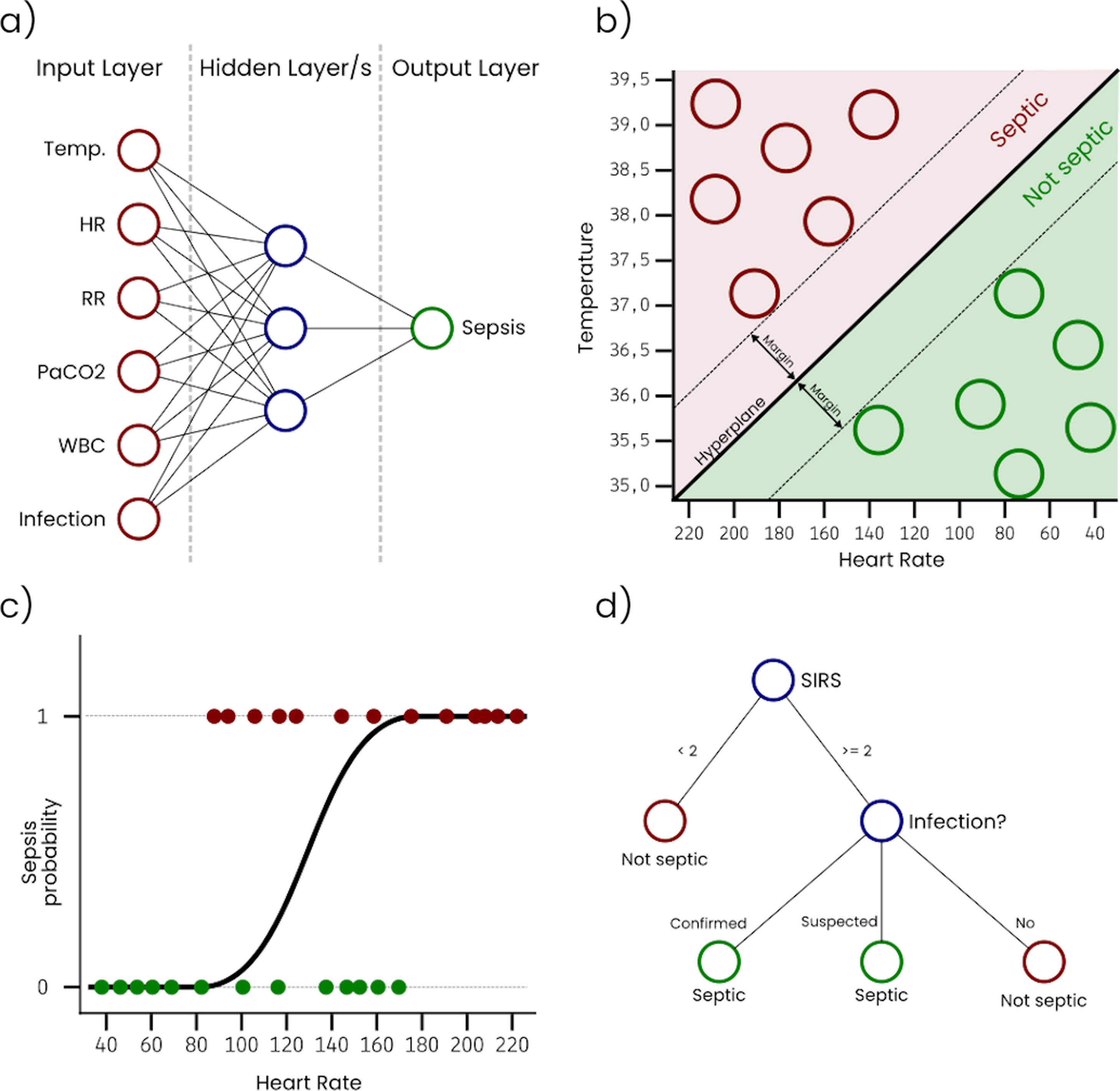
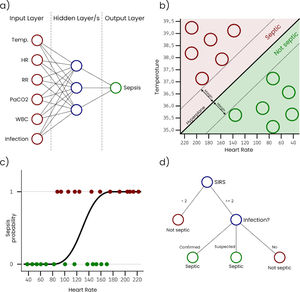
Artificial Neural Networks (ANN) are inspired in the way that neurons in a human or biological brain work. An ANN is an interconnected group of nodes (artificial neurons) separated into, at least, three layers: (i) input, that receives the sample data, (ii) hidden, that transforms the input values, and (iii) output, that provides the final prediction for each sample. However, it is very common to have more than one single hidden layer, as more layers increase the ability of the ANN to learn more complex problems. Fig. 1a represents a simple ANN with six input neurons, each one corresponding with a variable, one hidden layer with three neurons, and an output layer with a single neuron that outputs the sepsis probability. As can be seen, nodes on each layer are connected with all the nodes on the next layer. These connections are usually initialized with a random weight. The output layer is typically configured to have a node for each possible output condition, and the activation level of each of these nodes for a sample corresponds with the probability of the sample to belong to the condition associated with the node. During the training, the connection weights are adjusted, so that input values can be transformed into the expected output by successively applying mathematical transformations based on the connection weights. ANN classifiers generally have good performance, however, they are complex to configure, costly to train, and difficult to interpret.
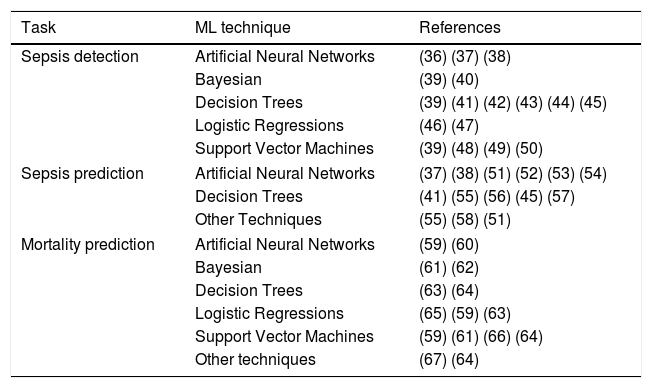
Support Vector Machines (SVM) treat each sample as a point in a n-dimensional space and try to find a hyperplane that separates samples from two different conditions. For this reason, SVM are considered binary classifiers, as they can only classify data with two possible outputs. However, there are several strategies to use SVM in problems with more than two possible outputs by combining several SVM classifiers. Fig. 1b represents a simple SVM with two input variables (i.e. temperature and heart rate). During the training, hyperplane parameters are adjusted to maximize the margin between the hyperplane and the samples with different outputs. SVM classifiers generally have good performance, however, they are complex to configure and trained classifiers are difficult to interpret.
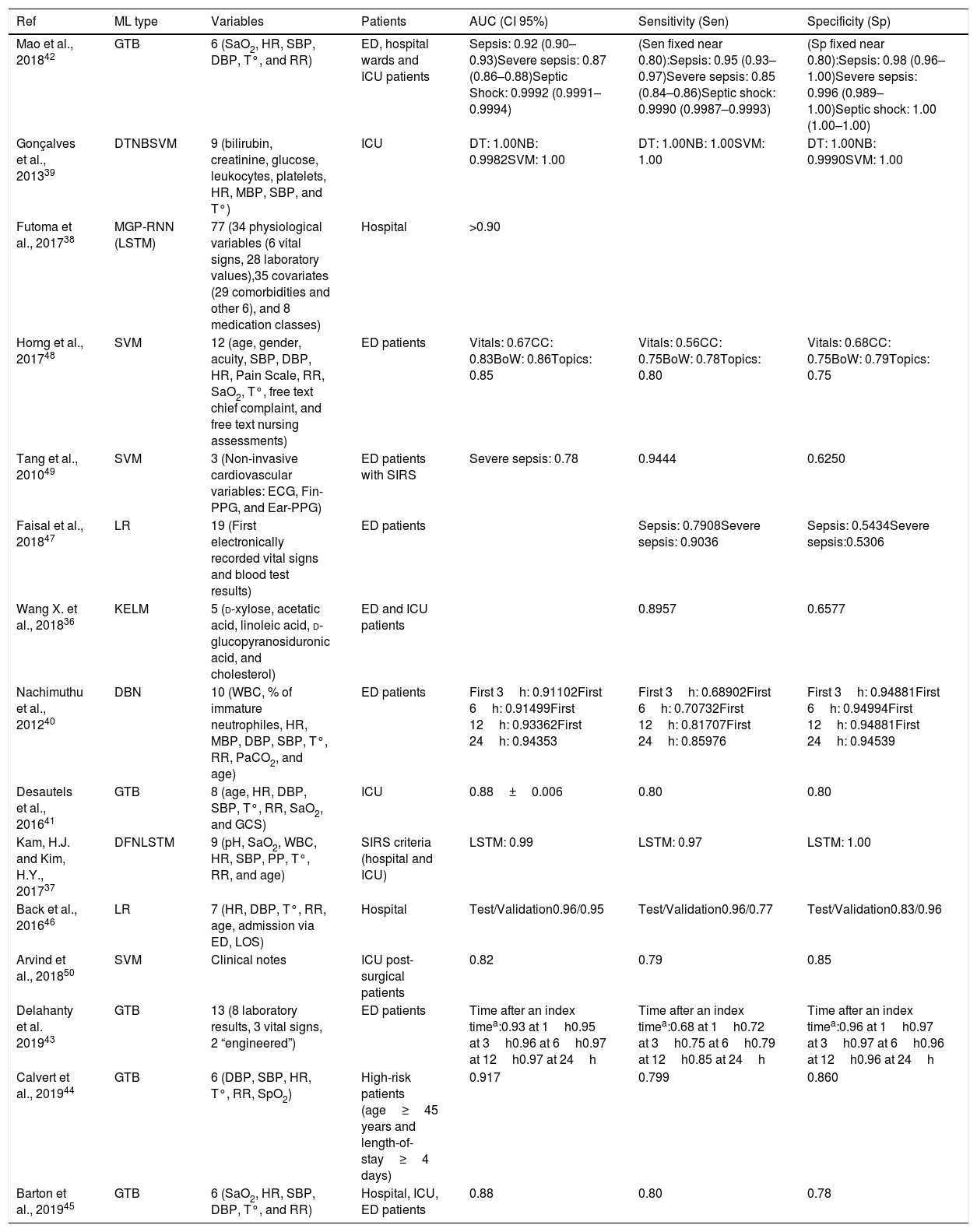
Logistic Regression (LR) takes its name from the logistic function on which it is based. Like SVM, LR algorithms are considered binary classifiers, but, in this case, the output will be close or equal to 0 for one condition and close or equal to 1 for the other condition. Fig. 1c represents a simple LR with a single input variable (i.e. heart rate). In this example, the sepsis probability increases with the heart rate, however this variable is not enough to predict sepsis, and some samples are misclassified. There is a generalized LR for multiple condition problems named Multinomial Logistic Regression. During the training process, the coefficients of the function used to generate the output are adjusted, so that the output value is as close as possible to 0 or 1 for each condition. Despite of its name, LR is used in classification problems. LR classifiers perform well on linear separable problems and are simple to configure and train. However, trained classifiers are difficult to interpret.
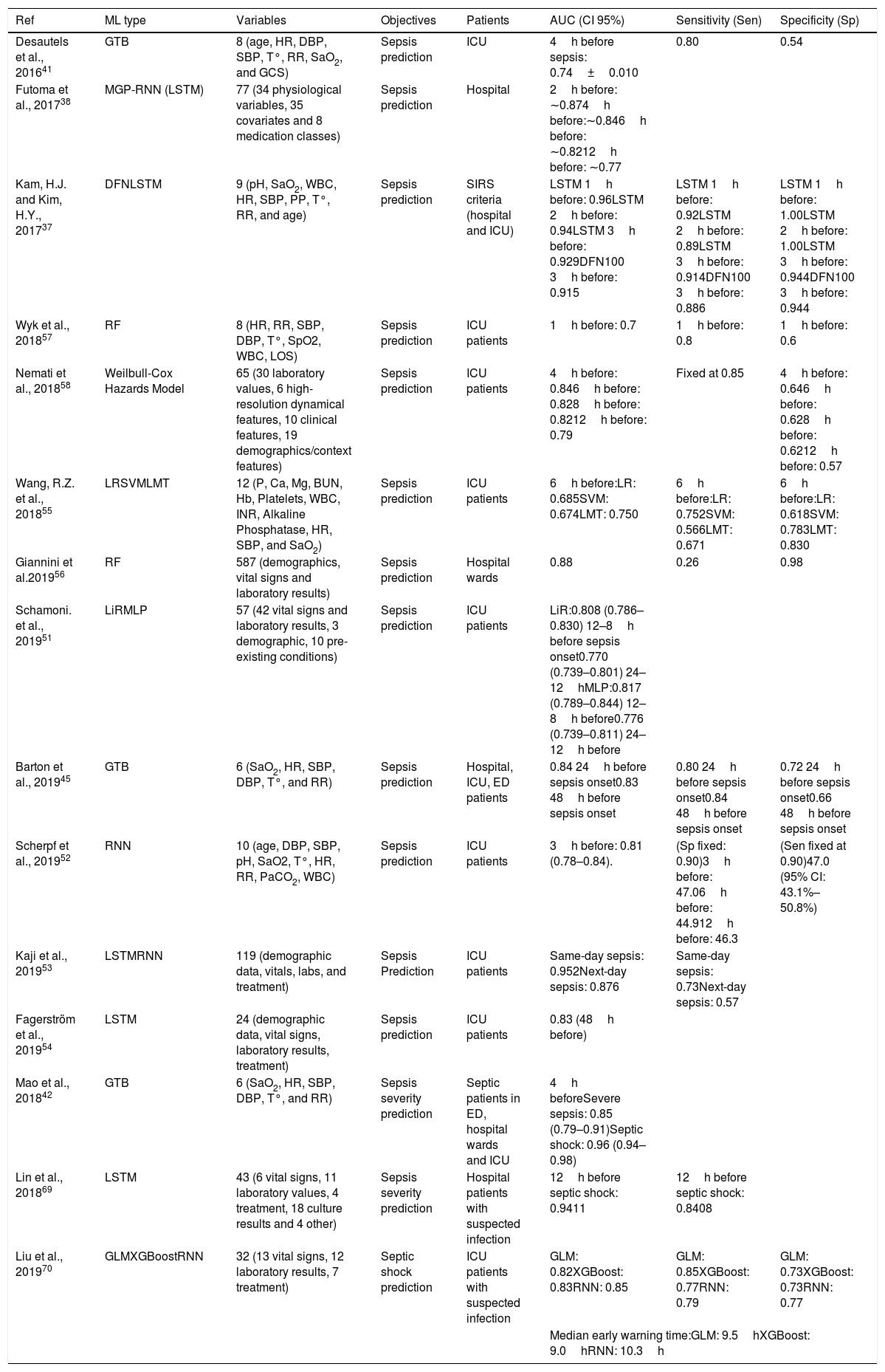
A Decision Tree (DT) algorithm has the internal structure of a tree graph, where each branch node evaluates a variable of the sample, each edge represents a possible outcome of the evaluation and each leaf node represents a possible outcome. Fig. 1d represents a simple DT that determines if a patient is septic following the sepsis-3 definition. The structure of the tree, including the number of nodes, the condition evaluated on each node and the value of each edge, is determined during the training process. A special type of DT is the Random Forest (RF), which is not strictly a tree but a set of DTs that use different variables of the samples. The final outcome of a RF is an average of the internal trees outputs, usually the mode, for classification, and the mean, for regression. RF classifiers generally have high performance, they are easy to configure and it is possible to get some useful information to interpret trained classifiers, such as the importance of the variables.
Bayesian algorithms, such as Naïve Bayes (NB) or Bayesian Network (BN), are based on Bayes’ theorem, which describes the probability of an event, based on prior knowledge of conditions that might be related to the event. This kind of algorithms use the training data to estimate the probabilities of each possible output based on the values of the variables of the training samples. NB assumes variable independence, which is not required by the BN. Bayesian algorithms are mainly used in text-based problems, although they can be used in many other domains. They are easy to configure and the trained classifiers are interpretable.
What are the key variables to build a clinical decision support system to assist in sepsis management?Taking into consideration the conducted review, and focusing the attention on the three main tasks identified in Table 1 (i.e. sepsis detection, sepsis prediction, and mortality prediction), we can see there are four different but complementary sources of information that are of utmost importance. First of all, a substantial number of papers mention vital signs (e.g. as heart rate (HR), respiratory rates (RR), temperature (Temp), blood pressure (BP), oxygen saturation (SaO2), etc.), and laboratory tests (e.g. renal and liver function, lactate level, coagulation profile, etc.). Additionally, certain patient characteristics (e.g. age, nationality, comorbidities) are taken into consideration to hypothesize different scenarios, whilst different severity scores such as SOFA (Sequential Organ Failure Assessment), APACHE (Acute Physiology and Chronic Health Evaluation), qSOFA (Quick Sequential Organ Failure Assessment), or MEWS (Modified Early Warning Scoring) are also used by multiple systems. Furthermore, some works emphasize the need to analyze complementary sources of information containing unstructured text such as nursing and medical notes, comments from different departments and personal of emergency with the goal of identifying clues about patients with sepsis.48
From a complementary perspective, some works indicate that a better management of clinical and administrative databases containing supplementary patient information is necessary to facilitate an automated prediction.38,46,58,63 In this line, the Electronic Health Records (EHR), already available in some centres and ICUs, eases the compilation, interchange, comparison, and effective use of medical information between different departments.
Can sepsis be automatically detected earlier through the use of ML techniques?Sepsis is a time-dependent syndrome, which implies that the sooner we start treatment, the better the prognosis.15,18 Although many efforts have been carried out in order to establish and update a precise definition of sepsis,5–7 the current identification of septic patients remains troublesome. An accurate sepsis detection system may change the way we approach its treatment.
Of the 34 works covered in the present study, 15 of them explicitly deal with the task of automatically detect sepsis.36–42,46–50 Main features of the most relevant works are commented hereunder.
Mao et al.42 developed a ML-based system (called InSight) that recognizes sepsis using only six common vital signs taken from the EHR of Emergency Department (ED), ICU or hospital wards patients (SaO2, HR, diastolic blood pressure (DBP), systolic blood pressure (SBP), Temp, and RR). The authors reported that InSight works correctly despite a significant amount of missing patient data. Faisal et al.47 developed a LR model to predict the risk of sepsis using only the first electronically recorded vital signs and adding first blood test results obtained following emergency medical admission, with good performance. Horng et al.48 demonstrated that the use of free text, in addition to vital signs and demographic information, allows for an increase in the ability of identifying infection at ED triage. Delahanty et al.43 and Barton et al.45 showed us that ML based models have a better performance than usually used (and recommended by clinical guidelines) scores, like qSOFA, NEWS or MEWS.
As we can see, most of ML-based sepsis detection systems use age, well-known vital signs (i.e. HR, RR, Temp, SBP, and DBP) and usual blood test results to build their models. Other authors use less common parameters. Arvind et al.50 showed us an interesting way of detecting sepsis using solely unstructured narrative discharge notes (Area Under Curve (AUC) 0.82). The work of Tang et al.49 is particular because of their use of electrocardiogram and finger and ear-lobe photoplethysmography signals to detect sepsis (AUC 0.78 for severe sepsis). There are, also, more experimental strategies, such as the one proposed by Wang et al.,36 who employed five biomarkers (d-xylose, Acetatic acid, Linoleic acid, d-glucopyranosiduronic acid, and cholesterol), reaching a sensibility of 0.896 and a specificity of 0.658.
To sum up, ML based model have a good performance to detect sepsis, even with a few vital signs obtained routinely, and without need for suspicion of sepsis, at ED admission or during hospital stay. Specific data and results of sepsis detection studies in Table 2.
Is sepsis prediction an achievable goal?As we have already explained, sepsis is a time-dependent syndrome, so we must detect it as fast as we can. Would it be possible even to predict that an infected patient is going to have a sepsis a few hours before its appearance or that a sepsis is going to deteriorate into septic shock? There are some approaches that expressly address this challenge.
Of the 34 works covered in the present study, 12 of them explicitly deal with the task of sepsis prediction and sepsis severity prediction. From a quantitative perspective, some studies were carried out using large datasets.39,41,50,58
Mao et al.42 built a model using only six vital signs able to predict evolution (4h before its appearance) to severe sepsis (AUC 0.85), and to septic shock (AUC 0.96) in septic patients in ED, hospital wards, and ICU. Using vital signs and combining them with other variables (laboratory results, treatment received, etc.), Lin et al.69 and Liu et al.70 achieved a higher prediction capacity, being able to predict evolution to severe sepsis (AUC 0.94) or septic shock (AUC 0.82) before severe sepsis (12h before) and septic shock (9h before) appearance.
It seems more interesting and challenging to predicting those non-septic patients admitted to hospital wards or ICU who will suffer from sepsis. Some authors have developed systems able to predict sepsis using just a few variables that are usually collected by the EHR (such as age, HR, RR, BP, and SaO2). Scherpf et al.,52 combining 10 variables (age, DBP, SBP, pH, SaO2, Temp, HR, RR, partial pressure of carbon dioxide (PaCO2), and white blood cell (WBC)) achieved an AUC of 0.81 to predict sepsis in ICU patients 3h before its appearance. Barton et al.,45 using just six variables (SaO2, HR, SBP, DBP, Temp, and RR), were able to predict sepsis development in hospital, ICU, and ED patients, with an advance of 24h (AUC 0.84) and 48hours (AUC 0.83).
Other more complex approaches, as Giannini's et al.,56 combined up to 587 variables to achieve an AUC of 0.88 to predict sepsis in hospital wards patients with one hour advance. As we can see, there are different systems that have demonstrated their ability to predict sepsis before its appearance, some of them based on a few variables usually recorded on EHR, with a good performance. Of course, accuracy increases closer to sepsis onset and when the system can have more dates of trends of each variable. Specific data and results of sepsis prediction in Table 3.
Can ML techniques be used to accurately predict sepsis-related mortality?Predicting sepsis-related mortality is important in order to both classify patients by their severity and identify those situations in which a more aggressive treatment may be necessary. Moreover, it could be useful for designing clinical trials, aiding in the selection of the target population. A tool that makes mortality prediction would be very useful for the selection of patients to be included in these studies.
Of the 34 works covered in the present study, 9 of them explicitly deal with the task of predicting sepsis-related mortality. There is a significant variability in the size of patient databases, ranging from large datasets to a few individuals under consideration.59 In most cases, prediction studies were carried out using smaller datasets when compared with the previous task.59,62
Both the ability to predict short-term and long-term mortality have been analyzed by different authors.
Short-term mortality prediction has been studied by Perng et al.60 They used 53 clinical variables, all of them obtained during ED patient stay, and achieved an AUC of 0.94 to predict mortality at 72h and of 0.92 at 28 days. Similar results (but no better) showed Taylor et al.63 with a more complex algorithm that combines more than 500 variables and achieves an AUC of 0.86 to predict sepsis related mortality at 28 days in ED patients.
Garcia-Gallo et al.67 developed a ML-based model for predicting 1-year mortality in critically ill patients diagnosed with sepsis, using the MIMIC-III critical care database.71 Reported results using a tree-based ensemble classifier outperformed those obtained by other traditional scoring systems (e.g. SAPS II, SOFA or OASIS), reaching an AUC of 0.804. In their model, they include 47 variables (including vital signs, laboratory results, comorbidities, and organ dysfunction).
Regarding the variables used, most of the proposed models use age, lactate, WBC count, and other well-known vital signs (as RR, Temp, and mean blood pressure) all of them easily accessible in different settings. From a complementary point of view, the work of Byrne et al.59 is especially interesting because mortality is predicted by using 939 peptides identified with LC-MS/MS (Liquid Chromatography with tandem Mass Spectrometry) in blood samples.
To sum up, different works have shown a relatively good performance predicting long and short-term sepsis related mortality, usually employing more variables than in previous tasks. Specific data and results of mortality prediction in Table 4.
Is it possible to effectively increase adherence to treatment guidelines and reduce sepsis-related mortality and/or associated costs using ML techniques?Despite the fact that it is difficult to assess how sepsis detection (and especially, sepsis prediction) will change sepsis treatment, in this work we have tried to elucidate if ML techniques could improve, by themselves, sepsis treatment. Due to the high mortality related to sepsis,8,10 increasing survival should be the goal when designing an intervention. However, sepsis also generates huge health resources consumption,13,14 so a parallel objective should be to reduce the cost per episode. In our review, we found that both objectives (i.e. clinical and economic) could be achieved through the effective use of ML techniques.
The AI Clinician,72 a computational model using reinforcement learning, developed from the analysis of treatment received by patients in the MIMIC-III, is able to dynamically suggest optimal treatments for adult patients with sepsis. The system has been able to identify optimal fluid and vasopressors management from suboptimal training examples. In an independent validation cohort, comprising patients from the eICU Collaborative Research Database,73 those who received the treatment that the AI Clinician would recommend had the lowest mortality rate. Authors suggest that this system could be used in a real environment, proposing a course of action for the septic patients in real-time. Although they do not expect to replace the physician, as the selection of the treatment strategy still would require their clinical judgement, they think that the system could provide additional insight about optimal decisions to increase the patient survival expectative.
There are, also, systems that are prospectively tested. The impact of the InSight system,42 described above, was assessed in two prospective studies. McCoy and Das74 showed how after deploying InSight (compared with pre-implantation period), in-hospital mortality rate decreased by 60.24%, sepsis-related hospital length of stay (LOS) decreased by 9.55%, and sepsis-related 30-day readmission rate decreased by 50.14%. The study was carried out with 1328 cases and showed how early intervention can reduce mortality and LOS, thereby decreasing the overall hospital cost. Furthermore, Shimabukuro et al.75 conducted a randomized clinical trial in two ICUs with the goal of calculating the average LOS and in-hospital mortality rate of two groups of patients (i.e. 67 experimental patients vs. 75 control patients). InSight obtained a decrease of the average LOS of 2.7 days between control and the experimental group (representing a 20.6% reduction). Additionally, the mortality rate showed a decrease of 58.0%.
Giannini et al.56 also tested their system in a pre-post trial, resulting just in an increase of lactate testing and intravenous fluids administration with a reduction in time to ICU-admission without impact on mortality or ICU LOS before ML system was implanted.
Discussion and conclusionsIn the future, application of ML systems to sepsis diagnosis and management could change our way of dealing with this pathology. First of all, criteria for sepsis diagnosis could change: until now, the simplicity of the definition was a priority, since sepsis can occur at any level of care and must be recognized by professionals not specialized in this field. This simplification has led us to the point that, nowadays, sepsis diagnosis is based on the presence of established organic dysfunction. A sepsis diagnosis tool based on ML could analyze a massive number of variables not affordable to us. This system, properly calibrated, would allow us to reach a more precise diagnosis, and even predict the appearance of sepsis, completely changing the management of this entity. Secondly, the development of clinical decision support systems (CDSS) with complex algorithms based on AI may improve adherence to accepted management recommendations, giving individualized treatment advice. Finally, employing Big Data and ML based algorithms could let us know a precise outcome for each specific patient.
These systems, although they do not replace human doctors, offer a series of advantages over them: they would allow almost all hospitals around the world to have a sepsis expert present for 24h 7 days a week, who does not get tired, and who always offers evidence-based treatment.
However, ML-based health care is far from ideal. We must also emphasize that there are serious difficulties for the development of intelligent systems for health care. First of all, not all ICUs and hospitals are using EHR today, something that is essential to apply a ML based system. On the other hand, as pointed out by Beam and Kohane,76 ML “is not a magic device that can spin data into gold” directly, as ML is a natural extension of statistics to deal with and take advantage of the huge amounts of data available nowadays, but a big human and scientific effort still is needed to let a machine learn in each specific scenario. In fact, one of the most important elements for ML, if not the most important, is data. ML needs large, experts-curated and, most importantly, labelled, datasets that should be extracted and properly processed. Even though such large datasets could be collected, they may be subject to biases.77 In this sense, the freely accessible to MIMIC-III critical care database is the most employed source of data for training sepsis-related models, instead of private datasets, showing that it is not easy to find. Although this database has an undoubted value and quality, it only contains ICU patients, where data is recorded very frequently. In fact, many ML efforts go to where data is available, most of times forgetting about its real clinical value.78 As an example, in the case of sepsis, it is obviously useful to detect or predict sepsis earlier in ICU or ED, using routine variables that are recorded with no need for “human” sepsis suspicion. However, what about doing this outside intensive vigilance services? A system predicting this event earlier in hospital wards could be of breakthrough value, but it would require continuous monitoring of the patients with real-time registration of the generated data into their EHR, which is not feasible. On the other hand, despite their high AUC to detect and even predict sepsis, it is very difficult to calibrate these systems, as increasing the specificity and sensitivity of them is usually at the expense of each other. Therefore, if it has a high sensitivity it is going to trigger many unnecessary alarms with the resulting fatigue, whereas if it has a high specificity, some patients are not going to be detected.
One argument against ML could be that some of the relationships stablished by these systems are not explainable from a physiopathological point of view,79 however, some authors like Eric Topol80 defend that black box procedures, whose action mechanism is unknown, are already accepted in medicine and, therefore, black box ML systems should be also accepted. Moreover, ML could provide us with a more deep understanding of sepsis, opening up new ways to deal with it.
Another key aspect of ML in health care is the rigorous evaluation of the proposed models. New ML-based systems for health care are proposed on a daily basis, but many of them are retrospective studies. Clinical prospective studies are more difficult to find and randomized controlled trials of AI systems are still an exception. These trials pose challenges for patient or physician-level randomization, since two different patient workflows should be used for the treatment and control groups, which can be perceived as a risk and could be not be finally authorized by providers.78 In the case of sepsis, the work of Shimabukuro et al.75 is virtually the unique case where a clinical trial has been carried out.
Other aspect to take into account is the road to the market, which is probably not going to be easy, as for traditional drugs.81
Finally, ethics plays a key-role in ML for health care, also in sepsis. A well-calibrated prognosis predictor on such critical condition could even raise dilemmas, such as: is it acceptable not to initiate or withdraw support measures in patients with a probability of dying above a certain threshold? Could the results be artifacted by our prejudices when it comes to treating real patients?
ML is a very promising tool to improve sepsis detection and management, but there is probably a long way in front of us and to go along it, we will need to work as a team with partners unknown until now, like AI experts.
Author contributionInitial concept and design: D. Glez-Peña, P. Vidal-Cortés.
Clinical support for the research questions: P. Vidal-Cortés, L. del Río Carbajo.
Literature review and discussion: N. Ocampo-Quintero, M. Reboiro-Jato, F. Fdez-Riverola.
Drafting of the manuscript: N. Ocampo-Quintero, L. del Río Carbajo, F. Fdez-Riverola,
Final approval: N. Ocampo-Quintero, P. Vidal-Cortés, L. del Río Carbajo, F. Fdez-Riverola, M. Reboiro-Jato, D. Glez-Peña
FundingThis work was partially supported by the Consellería de Educación, Universidades e Formación Profesional (Xunta de Galicia) under the scope of the strategic funding of ED431C2018/55-GRC Competitive Reference Group. N. Ocampo-Quintero is supported by a doctoral scholarship from National Council of Science and Technology (CONACYT) (member identification number: CVU 681045) from Mexico.
Conflict of interestsThe authors have no conflict of interest to disclose.
SING group thanks CITI (Centro de Investigación, Transferencia e Innovación) from University of Vigo for hosting its IT infrastructure.