Early diagnosis and treatment has an important impact on the morbidity and mortality of infections caused by multidrug-resistant bacteria. Multidrug-resistant gram-negative bacilli (MR-GNB) constitute the main current threat in hospitals and especially in intensive care units (ICU). The role of the microbiology laboratory is essential in providing a rapid and effective response. This review updates the microbiology laboratory procedures for the rapid detection of BGN-MR and its resistance determinants. The role of the laboratory in the surveillance and control of outbreaks caused by these bacteria, including typing techniques, is also studied. The importance of providing standardized resistance maps that allow knowing the epidemiological situation of the different units is emphasized. Finally, the importance of effective communication systems for the transmission of results and decision making in the management of patients infected by BGN-MR is reviewed.
Un diagnóstico y tratamiento precoz tiene un impacto importante en la morbilidad y mortalidad de las infecciones producidas por bacterias multirresistentes. Los Bacilos gramnegativos multirresistentes (BGN-MR) constituye la principal amenaza actual en los hospitales y muy especialmente en las unidades de cuidados intensivos (UCI). El papel del laboratorio de microbiología es esencial en dar una respuesta rápida y eficaz. En esta revisión se actualiza los procedimientos del laboratorio de Microbiología para la detección rápida de los BGN-MR y de sus determinantes de resistencia. También se estudia el papel del laboratorio en la vigilancia y control de brotes por estas bacterias, incluyendo las técnicas de tipificación. Se destaca la importancia de proporcionar mapas de resistencia normalizados que permita conocer la situación epidemiológica de las diferentes unidades. Finalmente se revisa la importancia de sistemas de comunicación eficaces para la transmisión de resultados y la toma de decisiones en el manejo de pacientes infectados por BGN-MR.
Antimicrobial resistance is a problem of public health worldwide. The number of infections due to multi-drug resistant bacterie is on the rise every year in numerous centers, not only hospital-acquired but also community-acquired.1 There are numerous pieces of evidence that confirm that an early and proper management of multi-drug resistant baceteria-related serious infections reduces morbidity, mortality, and length of stay.2 This is particularly relevant in intensive care units (ICU) where the risk factors for aquiring this type of infections is particularly relevant. Currently, the most significant infections are due to gram-negative bacteria—especially enterobacteriaceae—and particularly Klebsiella pneumoniae K. pneumoniae), extended-spectrum β-lactamase-producing Enterobacteriaceae (ESBL-E) and/or carbapenem-producing Enterobacteriaceae (CPE), Pseudomonas aeruginosa (P. aeruginosa), and Acinetobacter baumannii (A. baumannii), which are resistant to carbapenems. Also, these multi-drug resistant baceteria can belong to high-risk clones thus adding the capacity of spreading and colonizing patients or environmental surfaces to its existing multiresistant capabilities.3 These bacteria that are resistant to almost all beta-lactam antibiotics are also resistant to other antimicrobial families such as quinolonas or aminoglycosides. In some cases, there are few therapeutic alternatives to treat infections due to these microorganisms. Empirical broad-spectrum antimicrobial therapies can promote the development of resistance at the ICU setting. To avoid this, early data on identification, resistance mechanisms, and antimicrobial sensitivity of the infection causing microorganisms are needed. In this case, te role of the microbiology laboratory is essential. It should be able to provide information on etiology and antimicrobial sensitivity/resistance in the shortest time possible in such a way that targeted therapies can be used to improve the prognosis of patients with the smallest possible impact on the phenomenon of antimicrobial resistance.4,5 The methods and techniques used should be adapted to the epidemiological reality of each center. However, providing this information fast is particularly useful in units with severe infections or patients with severe baseline conditions as it occurs at the ICU setting. To carry out these activities, access to the microbiology laboratory on a 24/7 basis is essential. In this manuscript we will be reviewing the role of microbiology laboratories in the diagnosis and treatment of multiresistant gram-negative bacilli (MR-GNB), as well as in the monitorization and control of infections due to these microorganisms at the ICU setting
Diagnosis of infections due to MR-GNB. Recommendations for sample drawing and meaning of microbiological findingsInfections due to MR-GNB in critically ill patients admitted to the ICU are mainly associated with mechanical ventilation, catheter-related bacteremias, surgical site infections, and urinary tract infections. Still, all organs or tissues can be infected.6 These microorganisms initiatlly colonize the patient’s skin and mucosas. They are acquired from surfaces, the hands of the healthcare personnel or colonized surfaces at the ICU setting or else after the selection of antibiotics based on the patient’s intestinal flora. The patient’s colonization increases the chances of developing a novel infection due to these microrganisms.7,8 The case of CPE can affect up to 17% of colonized patients.9 The elevated degree of colonization of patients at the ICU setting complicates the correct interpretation of microbiological findings. The culture of MR-GNB in sterile samples like blood, pleural fluid or aspirates are indicative of infection almost 100% of the times. However, isolating respiratory samples, the urine of catheterized patients or samples from open wounds requires a clinical correlation to distinguish colonization from infection. In this sense, quantitative cultures of respiratory10 and urine11 samples improve specificity by using cut-off values for these samples (Table 1). Gram stain can be used in these samples and provide information suggestive of contamination, as well as for the early diagnosis of pneumonia.
Microbiological analysis of a culture of different samples.
| Type of sample | Cut-off value | Additional parameters that should be studied |
|---|---|---|
| Respiratory samples | ||
| Bronchoalveolar lavage | ≥104 CFU/mL | ≥5% cells with intracellular bacteria |
| Protected specimen brush | ≥103 CFU/mL | |
| Endotracheal aspirate | ≥106 CFU/mL | <1% of epithelial cells |
| Urine sample | ||
| Urine from vesical catheter | ≥100 CFU/mL | |
| Permanent catheter | ≥1000 CFU/mL | Leukocytes in urine |
The most widely used methods based on traditional cultures are (Table 2):
Technical characteristics for the rapid detection of resistance in MR-GNB.
| Relevant characteristic | Selective/chromogenic media | MALDI-TOF | Immunochromatography | Colori metric | Accelerate Pheno | rAST-EUCAST | Alfred 60 AST |
|---|---|---|---|---|---|---|---|
| Easy to use | +++ | ++ | +++ | +++ | ++ | +++ | ++ |
| Interpretation of results | Growth vs. no growth | Analysis of peaks in the spectrum | Visible lines in a membrane for each type of carbapenemase | Change of color | Interpretation using the S/R software | Inhibition areas around the discs after a 4−8 h incubation perior | Analysis of growh curves |
| Result time | 24 h | 4 h | 15 min | 30 min-2 h | 8 h | 4−6 h | |
| Cost | + | ++ | ++ | ++ | ++++ | + | +++ |
1. Selective culture means. They can be prepared at the lab or commercial means can be used. Some of the latter include colorless chromogenic substrates that, due to the specific enzymatic activity of each microorganism, give the colony an intense and specific color for presumptive identification purposes. These means should be used as the screening method. Also, the species and mechanism of resistance should always be confirmed with other methods.12,13
2. Method to detect carbapenemases based on spreading with carbapenemase-inhibitor discs, ESBL, and AmpC.14
MALDI-TOF is a mass spectrometry system to rapidly identify bacteria by analyzing proteins. Also, it can be used to detect carbapenemase activity through the detection of carbapenem hydrolysis.15,16 This technique can be cost-effective with the proper machine. However, it requires a special module for analysis, reactive preparation, trained personnel capable of carrying it out, and interpreting it. Reactives are often prepared at the lab, although there are commercial kits available like the MBT STAR-Carba (Bruker Daltonics) capable of detecting carbapenemases.17,18 This test, however, does not provide information on other possible mechanisms of resistance to carbapenems or identifies the type of carbapenemase. A new application of this system, still under experimentation, is the detection of plasmid associated proteins carrying carbapenemases.19
Immunochromatography is an easy technique that is also easy to read and interpret. It is based on the use of carbapenemase-type specific antibodies.
One of the most widely used methods is the NG-Test Carba 5 capable of detecting the main carbapenemases (KPC, OXA-48, NDM, IMP, VIM), although false positives have been reported (100% sensitivity, >95% specificity). Results are ready in just 15 min even with isolates from positive blood cultures.20,21 Recently, this technology has been adopted for the derection of CTX-M extended-spectrum beta-lactamases (ESBL).22
Rapid colorimetric methods can detect the presence of enzymes (carbapenemases, ESBL) through substrate hydrolysis that changes pH and color too. Currently, several commercially available tests are used like RAPIDEC CARBA NP (Biomerieux), β-CARBA (Bio-Rad), β-LACTA (Bio-Rad), and MAST PAcE (Mast Diagnostics). Some of these tests have a low sensitivity to detect OXA-type enzymes and in the detetion of rare carbapenemases.23,24
Finally, there are rapid antibiogram methods available that are not focused on the detection of a signle phenotype of resistance but allow us to obtain information from more antimicrobials.
Accelerate pheno system (Accelerate diagnostics)It is an automated identification system and antibiogram of positive blood samples in approximately 8 h. Through a morphokynetic analysis, the system analyzes phenotypic characteristics like size, shape, and rate of division of the bacteria that grow in microcolonies in the presence of antibiotics. Identification match sits at around 87.9%–100% while in the clinical category it goes as high as 91%.25 The main drawbacks are its cost, the fact that it only determines the sensitivity of the species included in its identification panel, and its lower performance in polymicrobial infections. Studies using sterile fluids in blood culture flasks have been conducted. However, its performance is lower due to the greater chances of including polymicrobial infections or infections due to microorganisms not included in the panel.25,26
Rapid antibiogram reading based on the EUCAST criteria (rAST-EUCAST)EUCAST has validated a cheap and easy-to-use method to conduct rapid antibiogram readings (4 h, 6 h or 8 h of incubation time) by using the spread method with discs after inoculating the plaques directly from a positive blood culture flask.27,28 It is a standard method that has only been validated for 8 different species (Escherichia coli, K. pneumoniae, P. aeruginosa, Staphylococcus aureus, Streptococcus pneumoniae, Enterococcus faecalis, Enterococcus faecium, and A. baumannii). This method requires the rapid identification of the microorganism for the selection of the antibiogram. One of its setbacks is that if final outcomes are not obtained after 8 h, then a conventional angiogram needs to be performed. Different studies have been conducted comparing rAST-EUCAST with other methods, and good match has been reported.29
Alfred 60 AST (Alifax)This system provides an angiogram directly from positive blood culture flasks in 4 h–6 h. The system measures the dispersion of light to detect bacterial growth in a liquid environment with antibiotics by estimating a percentage of growth inhibition compared to growth in an environment without the antibiotic. This method does not identify bacteria, which is why rapid previous identification is required. Comparative studies with other techniques confirm that match is around 90% and 95%.30,31
Currently, it is in the pipeline together with new techniques based on microfluids, biosensors, optics, electronics, and DNA amplification that will revolutionize the detection of antimicrobial resistance.32
Molecular techniquesThe detetion of MR-GNB at the microbiology laboratory requires the correct identification of the bacterial species and resistance determinants. The most widely used methods to identify GNB are based on the detection of proteins (proteomic methods), like MALDI-TOFF, that identifies correctly most pathogenic multiresistant bacteria although presents specificity issues when detecting the most relevant resistance mechanisms, which is relatively tiresome.33 There is a wide variety of molecular techniques to identify resistance determinants, and the most widely used ones are based on the detection of nucleic acids (NA) (genetic methods) with a high sensitivity and specificity. Also, they that can be used from cologne or direct sampling.34 The most widely used genetic molecular methods are based on gene amplification reactions and amplicone detection and sequencing. The amplification method most widely used in the detection of MR-GNB is polyemerase chain reaction (PCR). However, over the last few years, other faster and more cost-effective aternatives like the loop-isothermal amplification technique (eg, LAMP) are being used.35 There are 3 large groups of NA amplification techniques on real time: target amplification-based techniques (eg, PCR, NASBA, etc.), signal amplification-based techniques (eg, bDNA, hybrid capture), and probe amplification-based techniques (LCR).36 The most widely known amplification technique is PCR both in its conventional format (simple PCR, nested PCR, multiple PCR, RT-PCR) and in real time.35 Amplicon detection can be performed through ethidium bromide-stained agarose gel electrophoresis (conevtional PCR), through hybridation with marked specific fluorescent probes (eg, Taqman probes, FRET probes, molecular beacon probes, Scorpions probes, etc.) or using NA sequence-based techniques.33,35
Sequencing NA allows us to identify determinant-coding genes of antimicrobial resistance, regulate genes of such resistance determinants, as well as mobile genetic elements like plasmids, and transposons that spread or disseminate these resistance determinants.
NA sequences commonly used today are based on Sanger sequencing (also called ´chain termination enzymatic method´—population sequencing). However, over the last feew years, new technologies or platforms have been developed for massive sequencing (clone sequencing) or high-performance pruposes (next-generation sequencing) like Illumina, PacBio or Nanopore to sequence complete bacterial genomes.37,38 Massive sequencing provides us with useful information regarding typification (see molecular typification section below) or to study virulence genes of MR-GNB.37
The main advantages of molecular techniques compared to conventional techniques (culture, etc.) are that they are fast and have a greater analytical sensitivity, which is why some of them—like the LAMP technique—are used as rapid techniques for the detection of MR-GNB (point-of-care) since they are particularly useful when applied to syndromic panels because they facilitate the detection of multiple multiresistant pathogens, and mechanisms of antimicrobial resistance.39,40 Sepsis panels can identify a huge number of multirresistant bacteria and mechanisms of resistance, and carbapenemases from positive blood culture flasks or blood samples with EDTA (Table 3). Many of these techniques have been automatized and are available in the market, and allow us to work with many different samples at the same time, which facilitates technical work and benefits the ultimate results.
Identification systems of sepsis-producing pathogens and resistance determinants approved by the FDA.
| Characteristic | System or plataform | |||||
|---|---|---|---|---|---|---|
| BioFire FilmArray (Biomerieux) | Verigene (Luminex) | ePlex BCID (GenMark) | Unyvero (Curetis) | Sepsis Flow Chip Assay (Master) | Magicplex Sepsis (Seegene) | |
| Technology | PCR multiplex | PCR + Microarray | PCR + Microarray | PCR + Microarray | PCR + Microarray | Conventional PCR and RT-PCR |
| Clinical sample | ||||||
| Positive blood culture | X | X | X | X | X | |
| Direct blood | X | |||||
| Gram stain | X | X | ||||
| Response time (h) | 1 | 2.5 | 1.5 | 4 | 3 | 6 |
| Manual manipulation time (min) | <2 | 10 | <2 | <2 | NC | NC |
| Gram-negative pathogens found | 24 | 22 | 57 | 35 | 18 | 91 |
| 11 | GN panel (9) | GN panel (21) | GN panel (15) | 10 | 12 | |
| Gram-positive | 8 | GP panel: 13 | GP panel: 20 | GP panel: 11 | 7 | 73 |
| Fungi | Candida spp. (5) | Fungi panel (16) | Fungi panel (8) | C. albicans (1) | Fungi (6) | |
| M. tuberculosis | X | |||||
| No. of resistance determinants | 3 | 8 | 8 | 16 | 20 | 3 |
| mecA | X | X | X | X | X | X |
| VanA/VanB | X | X | X | X | X | X |
| CTX-M | X | X | X | X | X | |
| OXA-48 | X | X | X | |||
| KPC | X | X | X | X | X | |
| VIM | X | X | X | X | ||
| NDM | X | X | X | X | ||
| IMP | X | X | X | X | ||
The most relevant limitations of genomic molecular techniques are their high cost and the need, in some cases, for trained personnel experienced in this type of techniques and biocomputing tools to analyze the information obtained.
Antibiogram interpretation. Resistance characterization algorithmsIn the interpretive reading of an antibiogram (S/I/R) the quantitative results obtained from assessing the activity of an antimicrobial vs a microorganism is performed through standard methods. During the 1990s, the concept of interpretive reading of the antibiogram appeared for the first time.41 This concept involves a complete phenotypical analysis of the results obtained vs all antimicrobials to infer possible mechanisms of resistance based on how they express phenotypically.42,43 Intepretive reading is a complex task for microbiologists since, although there are automated systems available with expert rules, microbial resistance is in continuous evolution and knowledge needs to be updated on an ongoing basis. As an example of interpretive reading regarding MR-GNB it is essential to analyze beta-lactam antibiotics all together since herein lays the foundation for the detection of ESBLs and carbapenemases, that stand out for their special epidemiological significance.
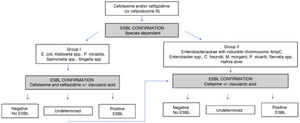
ESBL. In Enterobacteriaceae an almost infinite number of enzymes of this type (CTX-M, SHV, TEM) have been described. The properties of each enzyme, their levels of expression, and the presence of other possible additional mechanisms (altered membrane patency, active efflux pumps of antibiotics located in the membrane or presence of other betalactamases) facilitate a large diversity of phenotypes.44 The presence of ESBL should be suspected when the MIC of cefotaxime/ceftriaxone and/or ceftazidime is >1 mg/L (cefpodoxime >1 mg/L is a more sensitive but less specific indicator). Cefotaxime/ceftriaxone, and ceftazidime should always be tied in parallel since there can be differences in the MIC values based on the type of ESBL. MIC values for cefepime >4 mg/L, especially in E. coli and K. pneumoniae, confirm the presence of ESBL as the underlying mechanism. When after interpretive reading the presence of ESBL is suspected, it should be confirmed based on the in vivo inhibition of the activity of betalactamase by clavulanic acid.14Fig. 1 shows an example of the confirmation algorithm for the presence of ESBL according to EUCAST.
Carbapenemases. Same as it occurs with ESBL, multiple phenotypes can be observed due to the great variability of carbapenemases that has been reported.45 We should mention that carbapenem resistance can be mediated by the combination of different mechanisms of carbapenemases (eg, loss of porins in AmpC-hyperproducing strains). In carbapenemase-producing Enterobacteriaceae, the MIC values of carbapenems can be lower compared to the clinical cut-off values, which is why even lower cut-off values have been established (MIC >0.125 mg/L for meropenem or ertapenem, having the latter the highest sensitivity but lowest specificity) for the detection of carbapenemase. However, in P. aeruginosa we should bear in mind that most cases of carbapenem resistance are due to the combination of intrinsic or chromosomic mechanisms (alteration of OprD porin together with the hyperexpression of active efflux pump) although resistance to the remaining beta-lactam antibiotics, particularly the ceftolozane-tazobactam combination, requires ruling out the presence of a carbapenemase.46 In A. baumannnii, carbapenem resistance is mostly due to the presence of a OXA-type acquired carbapenemase, although other carbapenemases can be present too, which should be confirmed through genotypical methods.47
Molecular typification methodsMolecular typification techniques are useful for the monitoring and control of outbreaks due to MR-GNB because they allow us to know clonality among different isolates, identify reservories, and determine possible routes of transmission. The leading molecular typification techniques are based on pulsed field gel electrophoresis (PFGE), NA amplification through PCR, and NA sequencing.48,49 PFGE is the molecular typification techniques par excellence since it has high discrimination capabilities.50 These are some of the most important limitations regarding PFGE: high cost of the machines, difficult to use, difficulty differentiating some types of pulse with very close DNA strands, and the fact that the genomes of some bacterial strains are not digested with the restriction enymes used. Although there are several PCR-based typification techniques (Table 4), the most widely used is the PCR of repeated DNA sequences (REP-PCR).48,49 These techniques are less expensive, easier-to-us (some of them are semiautomatic and sold as Diversilab from bioMérieux), and often generate patterns of bands that are relatively easier to interpret compared to PFGE. The discriminatory capabilities of PCR techniques can be lower compared to PFGE, especially in some Enterobacteriaceae like E. coli and K. pmeumoniae. Both PFGE and PCR techniques need to be optimized or standardized at the laboratory to guarantee good reproducibility. To study small outbreaks of short evolution while trying to obtain rapid information, the use of a PCR technique would be advised in some cases. To study more complex outbreaks of longer duration in time while trying to know the evolution or dynamics of clones with the passing of time, as well as their geographical distribution, the use of PFGE is more cost-effective.
Most significant characteristics of some molecular typification methods of BGN MDR.
| Characteristic | PFGE | Rep-PCR | MLST | SNP |
|---|---|---|---|---|
| Reproducibility | Excellent | Buena-Excellent | Excellent | Excellent |
| Discriminatory capabilities | Excellent | Moderate-Excellent | Moderate | Excellent |
| Easiness interpreting data | Moderate | Moderate-Good | Good | Good |
| Easy-to-use | Moderate | Good | Moderate | Moderate |
| High performance | No | Yes | Yes | Yes |
| Cost | Moderate | Low-moderate | Moderate | Moderate-High |
| Time needed (days) | 3 | 1 | 3–4 | 5–10 |
NA sequencing-based typification methods can be based on the partial sequencing of some genes through Sanger sequencing or else through complete genome sequencing using high-performance massive sequencing techniques.51 The most representative example of Sanger sequencing applied to molecular typing is MLST, which is based on the obtention of sequence types (ST) through the partial amplification and sequencing of 6–7 preserved genes.52 This methodology has lower discriminatory power compared to PFGE, which is why it is often used in global epidemiological studies or to analyze the population structure of microorganisms. Massive sequencing (NGS) can be used for typification purposes based on the sequencing of 6–7 genes (MLST) or several thousands of genes (core genome ST; cgST, and whole genome ST; wgST) which allows us, in certain bacterial species like K. pneumoniae, to differentiate different linages within the same clon or ST.51 NGS techniques are especially useful due to their high discriminatory capabilities when changes are analyzed at the level of each nucleotide (SNP) since, in this case, small changes among different highly interrelated transmission chains can be identified providing us with accurate information on how outbreaks originate and new transmission chains are generated from the index case.53
Cumulative report on antimicrobial sensitivityThese reports are very important to know the time tendencies of sensitivity of the main antimicrobials of clinical use, as well as to select the right empirical therapies. They are an essential tool for the work of PROA teams, and for the development of clinical practice guidelines on the management of local empirical treatment.54 Most centers follow the EUCAST method for its development.55 As a general rule:
- •
Reports often come out on a yearly basis.
- •
They should include the antimicrobials that are detailed in the usual result reports.
- •
Usually, the S and I categories are combined.
- •
The percentages of the mechanisms of resistance of epidemiological interest should be reported (eg, carbapenemases, ESBL, etc.).
- •
They should only include isolates of clinical samples previously validaded by a clinical microbiologist.
- •
They should include one isolate per patient only (the first one).
- •
Results should be expressed as percentages, at bacterial species level, and only in the presence of ≥30 isolates.
The routine clinical practice here is to distinguish the results of some special services or units. The rates of sensitivity to antimicrobials at the ICU setting can be lower compared to the overall hospital, which is why differentiating them is very important regardin the decision-making process in these units.56 One of the main limitations, depending on the size of the unit, is that sometimes the minimum number of 30 isolates cannot be achieved. In these cases, percentages should not be informed, but if they are, the relative value should be provided.
Design of monitorization measuresCurrently, there is an ongoing debate on the utility of systematic studies of carriers as an additional tool in nosocomial transmission control programs of MR-GNB.57 Active monitorization of ESBL-E carriers has not proven capable of reducing transmission. However, it is associated with a high negative predictive value.58 Therefore, the systematic monitorization of ESBL-E would be ill-advised here except for outbreak situations with the sole purpose of sizing these situations. A different story is to use the results of epidemiological monitoring cultures to select empirical therapies of immunocompromised patients at the ICU setting.59 With this goal in mind, it would make sense to monitor both ESBL-E and the remaining MR-GNB.60 A different story is to use the results of epidemiological monitoring cultures to select the empirical treatment, a controlversial issue due to the limited value of this type of samples to predict the etiology of infections in patients at the ICU setting.59,60 Regarding monitorization of other MR-GNB like A. baumannii, P. aeruginosa or CPE no clear evidence exists to this date. Therefore, most expert consensuses still recommend monitorization in patients admitted to this type of units.
Once admitted to the intensive care unit patients are exposed to environmental reservories like contaminated surfaces and the bacterial biolayers of siphons.61,62 This justifies monitorization while the patient remains hospitalized. However, there is no data available on which the optimal length of stay really is. In the presence of an outbreak and in situations of hospital discharge or extended endemicity it is also important to pay attention to environmental samples since they can detect reservories or cleaning or decontamination flaws. One of the most important reservories are the sink siphons at the ICU setting, which have proven to be the source of many outbreaks in these units.63 However, outside the aforementioned settings systematic sampling is ill-advised since these aquatic systems are often colonized without significant repercussions reported in the pressure of colonization.64 Another important aspect here is the monitorization of semicritical devices like endoscopies65 or non-critical devices that are close to the patient like anti-scarring mattresses or tables.66 If detachable bronchoscopies are not used, the microbiological control of cleaning and the reprocessing of flexible bronchoscopes should be included in the hospital general protocol regarding the monitorization of cleaning and disinfection.
Communications strategiesAll the steps we have talked about here need a good communication system with ICU representatives and nosocomial infection teams.67 No rapid and truthful result has validity in itself unless there is a clinician behind it receiving this information immediately and making the right decisions. Currently, laboratory computer systems provide results in real time including information on the patient’s electronic health record and establishing alerts to inform the clinician on relevant information immediately. The use of alerts and smartphones has proven effective. In any case, oral or on-site communication from microbiologist to ICU clinicians is still an effective tool for the transfer of relevant information and discussion of other diagnostic or therapeutic measures that should be taken. Periodic meetings among expert clinicians in infectious diseases, intensivists, and microbiologis should be encouraged to discuss the most relevant cases and see what additional diagnostic or therapeutic measures should be taken. Having meetings, at least once a year, where clinicians can talk about their diagnostic needs at the microbiology laboratory is important as well. Finally, collaboration with intensive care specialists and microbiologists in the nosocomial infection field in each center stands out as an important part of this process.
Microbiology laboratories, in collaboration with PROA teams, should decide what information they should be providing and when. This is particularly important at the ICU setting.68 The detection and information of resistance determinants is controversial because international agencies recommend providing information with epidemiological purposes only. Currently, there are several computer systems for diagnostic back-up and PROA programs including diagnostic data, resistance data, and, sometimes, the use of antimicrobials to facilitate the decision-making process.69
Inter-center communication is important, especially if patients with MR-GNB from other health centers are recived to identify patients with MR-GNB and minimize the chances of new outbreaks.
Conflicts of interestNone reported.
Please cite this article as: López-Hernández I, López-Cerero L, Fernández-Cuenca F, Pascual Á, El papel del laboratorio de microbiología en el diagnóstico de infecciones por bacilos gramnegativos multirresistentes. Importancia de la determinación de mecanismos de resistencias, Medicina Intensiva. 2022;46:455–464.